|
The Genome Taxonomy Database (GTDB; https://gtdb.ecogenomic.org/) is an initiative to establish a standardised microbial taxonomy based on genome phylogeny. |
|
The genomes used to build this phylogeny are obtained from RefSeq and Genbank. This dataset includes an increasing number of microbial dark matter genomes obtained from metagenome-assembled genomes (MAGs) and single amplified genomes (SAGs), facilitating an improved genomic representation of the microbial diversity. Of course, all genomes are independently quality controlled using CheckM before inclusion in GTDB. The archaeal GTDB taxonomy is based on genome trees inferred with IQ-TREE from a concatenated set of 122 marker proteins. Taxonomic ranks are normalised using relative evolutionary divergence (RED) and the resulting taxonomy is manually curated. The current GTDB release includes over 250,000 bacterial and over 4,000 archaeal genomes, representing over 45,000 and 2,000 species cluster, respectively. The archaeal GTDB taxonomy is publicly available at the GTDB website (https://gtdb.ecogenomic.org/), and we invite community engagement and feedback on the taxonomy through an online forum (https://forum.gtdb.ecogenomic.org).
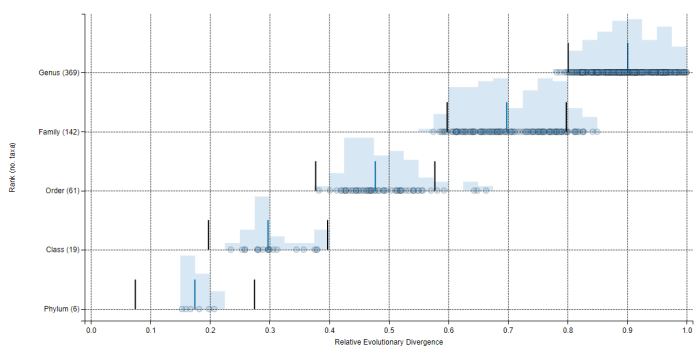
The relative evolutionary divergence (RED) of archaeal taxa at each taxonomic rank from phylum to genus in GTDB Release 06-RS202. RED values provide an operational approximation of relative time with extant taxa existing in the present (RED=1), the last common ancestor occurring at a fixed time in the past (RED=0), and internal nodes being linearly interpolated between these values according to lineage-specific rates of evolution. RED intervals for normalizing taxa at taxonomic ranks were operationally defined as the median RED value (indicated by a blue bar) at each rank ±0.1 (indicated by grey bars). Note that RED values are analysis-specific and should not be used as absolute values for comparison between studies. |